Another crucial aspect of these R&D efforts to inform antiviral drug design is accessibility of information. Lightstone points out, “We wanted to use DOE resources efficiently so the public wouldn’t have to repeat this work themselves.”
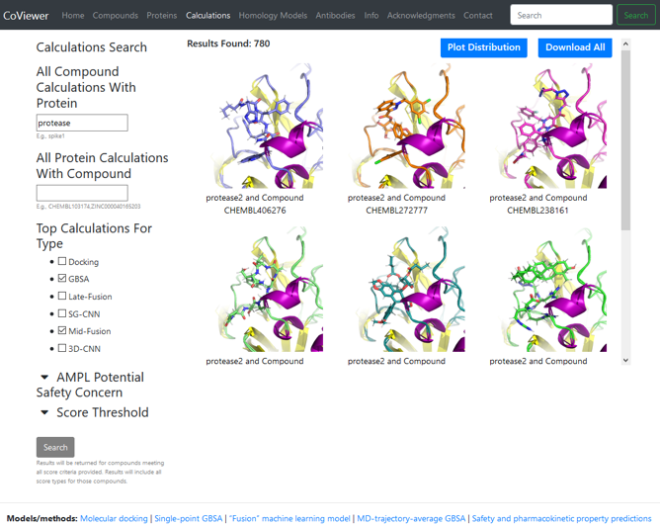
Much of the massive database of virtual screening results, chemical and protein structures, binding scores, and designed synthetic antibodies is available in an online data portal (covid19drugscreen.llnl.gov). Researchers anywhere can browse, filter, query, and download the data. Nicknamed CoViewer, the portal also provides details about the team’s research methods, ML models, calculations, and predictions. Torres explains, “CoViewer gives researchers a way to access this data and run their own algorithms on it.” (See Research Meets DevOps below.)
CoViewer debuted less than two months after the CARES Act was signed into law. The first batch of data included predicted antibody sequences, models of viral proteins from the viral genome, small molecule compounds from public sources, viral protein targets, and binding scores. The portal’s offerings have expanded in the past year with two additional data releases, and a third is expected to contain experimental results. “We’re scaling up the portal to include the full set of compounds that we’ve analyzed,” says Torres. Nearly 900 users visited CoViewer in the first half of 2021.
Just as pre-pandemic tools and technologies paved the way for these R&D activities, the latter will continue to advance LLNL’s biosecurity mission with applicability to other domains. For example, Torres states, “We’re continuing our collaboration with the American Heart Association that focuses on human proteins. It’s a similar but even larger dataset. We’ve learned a lot from the COVID-19 data about how to scale up for different types of workflows.” The team is also leveraging a graph-based dataset from the University of California, San Francisco, to improve predictions about drug efficacy.
Research Meets DevOps
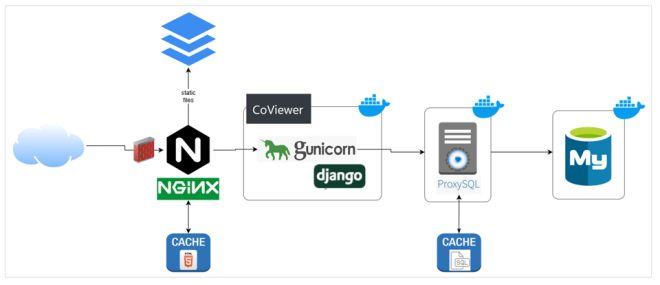
Computing’s 1.4-petabyte unclassified Green Data Oasis (GDO) server supports large-scale data storage and sharing with external collaborators. It works in tandem with LC’s commodity clusters like Catalyst and petascale machines like Lassen. In the spring of 2020, a portion of CARES Act funding was allocated to GDO to host the CoViewer data portal. Upgrades included additional storage, faster performance to handle data downloads and queries, and improved bandwidth and networking connectivity.
Working closely with the bioinformatics team, LC’s Workflow Enablement Group (WEG) facilitated the CoViewer deployment. “We provided the data and a container with our applications. Then WEG set up the host, moved the data, and managed the container,” explains Marisa Torres, emphasizing that a considerable challenge was determining how to search so much data. WEG web architect Thomas Mendoza adds, “Our expertise includes systems, containers, security, and web. We fit the bill for the deployment.”
As the project was under a significant time crunch, WEG staff prioritized features of the CoViewer web application and environment. Provisioning space on the GDO meant understanding data requirements, preparing firewall changes, and creating a development environment. “We needed to move quickly, but security was not an area for compromise. We leveraged our combined team expertise to ensure CoViewer would be secure, functional, and reliable,” states Mendoza. This effort accounted for rate limiting, logging, monitoring, and cyber security scans—everything to prepare the application for Internet-scale use.
The collaboration worked at a blistering pace—remotely—and launched the CoViewer in May 2020. “The GDO team performed miracles in standing everything up quickly,” says Lightstone. Now, more than a year later, the application remains low maintenance and spotlights Computing’s ability to optimize processes that accelerate web development and reduce overhead.